WSE-7066 EzRun MOPS non-SDS

Purpose and Application
-
Electrophoresis buffer for nucleic acid separation using polyacrylamide gels (PAG)
-
Can also be used for agarose gel preparation and electrophoresis of nucleic acids
-
Suitable for protein electrophoresis in Native PAGE / BN PAGE (MOPS-based buffer)
-
Compatible with Bis-Tris gels for protein Native PAGE / BN PAGE
Features
-
Expands the separation range on the low-molecular side (from 10 bp) simply by replacing the buffer
-
Enables shorter electrophoresis time
-
Sterilized and stable for 1 year at room temperature
-
Convenient small-volume type for easy handling
Data
Differences in Electrophoresis Buffers — EzRun MOPS non-SDS vs. EzRun TG

The left figure compares the electrophoretic patterns and mobilities of DNA using ready-made gels of the same concentration as self-prepared TBE gels, with different electrophoresis buffers. It demonstrates that WSE-7066 EzRun MOPS non-SDS (left) is more suitable for separating low-molecular DNA, while WSE-7055 EzRun TG (right) is better suited for high-molecular DNA. By selecting the appropriate electrophoresis buffer, it is possible to adjust the separation range even when using polyacrylamide gels of the same concentration.
Band Mobility Depending on Gel Concentration and Electrophoresis Buffer

The left figure shows the separation of a 20 bp DNA ladder (left) and a 100 bp DNA ladder (right) using 5%, 10%, and 15% polyacrylamide gels (e-PAGEL). Electrophoresis buffers were WSE-7066 EzRun MOPS non-SDS (M) or WSE-7055 EzRun TG (G). After electrophoresis, gels were stained with WSE-7130 EzFluoroStain DNA, excited with CyanoView, and imaged using the WSE-5400 Printgraph Classic.
Results demonstrate that WSE-7066 EzRun MOPS non-SDS produces lower band mobility compared to WSE-7055 EzRun TG. Even with gels of the same concentration, switching to WSE-7066 expands the separation range on the low-molecular side.
Ideal for Separating 10–20 bp DNA Fragments!

The figure on the left shows DNA separation using a 15% polyacrylamide gel (EHR-T15L) after mixing a 20 bp DNA ladder and DNA fragments of the indicated sizes (two 22 bp bands differ in sequence) with WSE-7040 EzApply DNA. WSE-7066 EzRun MOPS non-SDS (left) or WSE-7055 EzRun TG (right) was used as the electrophoresis buffer. After electrophoresis, gels were stained with WSE-7130 EzFluoroStain DNA, excited with CyanoView, and imaged using the WSE-5400 Printgraph Classic.
These results demonstrate that WSE-7066 EzRun MOPS non-SDS allows clear separation of DNA fragments in the 10–20 bp range according to molecular weight, with even 1 bp differences distinctly resolved. In contrast, using WSE-7055 EzRun TG results in bands separated at nearly the same positions with only minor differences. By simply switching the buffer to WSE-7066 EzRun MOPS non-SDS while using standard ready-made gels, DNA fragments can be efficiently separated without denaturation by urea or other reagents.
Native PAGE of Proteins with EzRun MOPS non-SDS
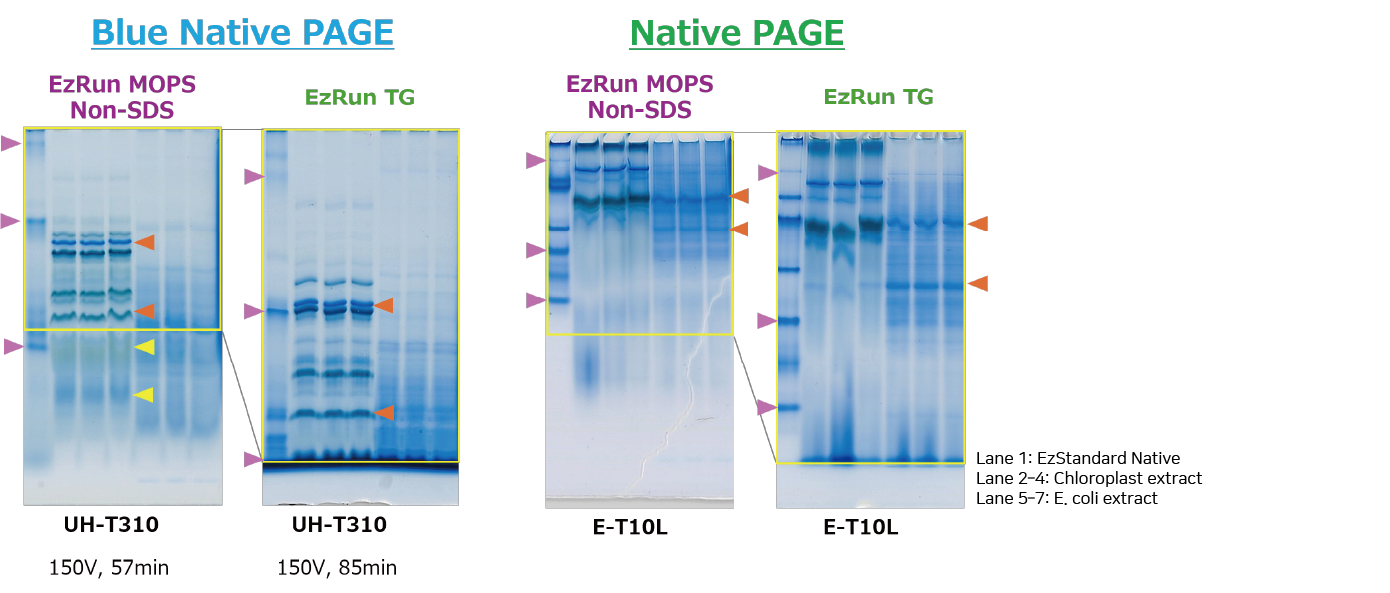
The figure shows the separation of chloroplast-derived proteins (extracted with WSE-7424 EzProteoLysis Native) and E. coli–derived proteins (extracted with WSE-7423 EzBactYeast Crusher), mixed with WSE-7011 EzApply Native, and applied directly to ready-made gels (u-PAGEL H 3–10% and e-PAGEL 10%). Samples were separated by Blue Native PAGE (left) or Native PAGE (right). WSE-7066 EzRun MOPS non-SDS (left) or WSE-7055 EzRun TG (right) was used as the buffer, with WSE-7067 EzBlueNative Additive added at 1/100 volume only to the Blue Native PAGE cathode buffer.
Results indicate that WSE-7066 EzRun MOPS non-SDS provides lower band mobility than WSE-7055 EzRun TG. Moreover, adding WSE-7067 EzBlueNative Additive enables simple Blue Native PAGE, while also achieving clear band separation in Native PAGE.
Blue Native PAGE of Bis-Tris Gels with EzRun MOPS non-SDS

The figure shows the separation of chloroplast-derived proteins (extracted with WSE-7424 EzProteoLysis Native) and E. coli–derived proteins (extracted with WSE-7423 EzBactYeast Crusher), mixed with WSE-7011 EzApply Native, and applied directly to ready-made gels (commercial Bis-Tris 3–12% gels and ATTO u-PAGEL H 3–14% gels). WSE-7066 EzRun MOPS non-SDS was used as the buffer, with WSE-7067 EzBlueNative Additive added at 1/100 volume only to the cathode buffer.
These results demonstrate that WSE-7066 EzRun MOPS non-SDS can be used not only with Tris/Tris-Glycine gels but also with Bis-Tris gels without interfering with electrophoresis. Furthermore, adding WSE-7067 EzBlueNative Additive enables simple and convenient Blue Native PAGE.
ATTO Native Electrophoresis Buffer Features
| WSE-7056 EzRun ClearNative | WSE-7057 EzRun BlueNative | WSE-7055 EzRun TG | WSE-7066 EzRun MOPS non-SDS |
|
|---|---|---|---|---|
| Protein denaturation | Non-denaturing (CN-PAGE) | Non-denaturing (BN-PAGE) | Non-denaturing (Native PAGE) | Non-denaturing (Native PAGE) |
| DNA electrophoresis | Not applicable | Not applicable | Compatible with Tris/Glycine agarose gels (Not for RNA) | Compatible with Tris/Glycine agarose gels (Not for RNA) |
| Electrode buffer system | Tris-HCl, Tris/Glycine, amino acid buffers | Tris-HCl, Tris/Glycine, amino acid buffers | Tris-HCl, Tris/Glycine, amino acid buffers | Tris-HCl–based, amino acid buffers |
| Features | Micelles of anionic surfactants contained in the running buffer bind to proteins, increasing their solubility and imparting a net negative charge to the entire complex. This allows electrophoretic separation according to molecular weight, independent of the isoelectric point (pI). The band resolution is reported to be higher than that of BN-PAGE. | When large amounts of negatively charged Coomassie Brilliant Blue (CBB) bind to proteins, their solubility is enhanced and the entire complex becomes negatively charged, enabling electrophoretic separation regardless of pI. Hydrophobic proteins bound to CBB remain stable without aggregation and are not dissociated by surfactants. In contrast, basic water-soluble proteins that do not bind CBB cannot be separated by this method. | Protein migration strongly depends on protein charge and isoelectric point. Negatively charged proteins (acidic proteins with a pI lower than the gel environment) migrate toward the anode, whereas positively charged proteins (basic proteins with a pI higher than the gel environment) migrate toward the cathode and may disappear. In some cases, proteins may precipitate during electrophoresis, resulting in smeared band patterns. | This system can be used for native electrophoresis of both proteins and DNA. In polyacrylamide gel electrophoresis, the mobility of both protein and DNA bands is increased, allowing clearer separation of low–molecular-weight bands. In particular, DNA fragments of 10–20 bp, which are typically difficult to resolve, can be clearly separated even on 15% gels. |
| Running Buffer / Gel Color | Clear | Dark blue (derived from CBB) | Clear | Clear |
| In-gel Enzymatic Activity Assay | Since protein denaturation is minimized, enzymatic activity is preserved. It is widely used for measuring the activities of OXPHOS, ATP synthase, NADPH-related enzymes, HRP, and various other enzymes. | Enzymatic activity may be inhibited due to binding of CBB. In addition, staining of the gel and proteins may interfere with the detection of enzymatic activity. | As protein denaturation does not occur, enzymatic activity is preserved. However, precipitation or reverse migration may occur during electrophoresis, sometimes preventing clear band separation. | Since protein denaturation does not occur, enzymatic activity is preserved. |
| ◎ | △ | ○ | ○ | |
| In-gel Fluorescence Assay | Since protein denaturation is minimized, fluorescence reactions are not inhibited, and absorption of fluorescent signals does not occur due to the clear background. It is used for analysis of Cy-dye–labeled proteins, GFP, YFP, and other fluorescent proteins. | Fluorescence detection may be inhibited because CBB binds to proteins. In addition, strong blue staining of the gel and proteins interferes with fluorescence detection by absorption, and fluorescence quenching of approximately 90–95% has been reported. | Since protein denaturation is minimized, fluorescence reactions are not inhibited, and absorption of fluorescent signals does not occur due to the clear background. However, precipitation or reverse migration may occur during electrophoresis, sometimes preventing clear band separation. | Since protein denaturation is minimized, fluorescence reactions are not inhibited, and absorption of fluorescent signals does not occur due to the clear background. |
| ◎ | △ | ○ | ○ | |
| Protein complex analysis | Because a small amount of an anionic surfactant is included, unstable interactions between subunits may dissociate during electrophoresis. As a result, some protein complexes may not be separated at their intact size. | Since dissociation of complexes does not occur during electrophoresis, complexes can be separated easily even in small amounts. This method shows high correlation with chromatography (≥90%) and is widely used for analysis of OXPHOS, ATP synthase, GPCRs, membrane proteins, and other protein complexes. | Although complex dissociation does not occur during electrophoresis, precipitation or reverse migration may occur, sometimes preventing clear band separation. | Complex dissociation does not occur during electrophoresis and this system is more suitable for separation of low–molecular-weight complexes than high–molecular-weight complexes. |
| △ | ◎ | △ | △ |
ATTO Electrophoresis Buffer Selection Guide
| Application | Running Buffer | Gel Buffer | Applicable Gels | Features | ||||
|---|---|---|---|---|---|---|---|---|
| Target | Protein | DNA | DNA | |||||
| Method | SDS-PAGE | Native-PAGE | DNA-PAGE | DNA-PAGE | Hand made Gels | ATTO Precast Gels | Agarose Gel | |
| AE-1411 EzRun | 〇 | - | - | - | 〇 Tris-based and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | - | A general-purpose Tris/Glycine/SDS running buffer compliant with the Laemmli method. Low cost; supplied as a powder for long-term storage. |
| AE-1412 EzRun C+ | 〇 | - | - | - | 〇 Tris-based and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | - | Contains reducing agents to suppress band broadening caused by re-oxidation during electrophoresis, enabling sharp band separation. Supplied as a powder. |
| AE-1415 EzRun T | 〇 | - | - | - | 〇 Tricine-based polyacrylamide gels | 〇 p-PAGEL, cp-PAGEL Neo | - | Tris/Tricine/SDS buffer for Tricine PAGE, suitable for separation of peptides and low–molecular-weight proteins. |
| WSE-7050 EzRun TAE | - | - | 〇 | 〇 | 〇 | Gel buffer / running buffer mainly used for DNA separation on agarose gels. | ||
| WSE-7051 EzRun TBE | - | - | 〇 | 〇 | 〇 TBE-based, Tris-based, and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | 〇 | Gel buffer / running buffer mainly used for DNA separation. Can be used with both agarose gels and polyacrylamide gels. |
| WSE-7055 EzRun TG | - | 〇 | 〇 | 〇 | 〇 Tris-based and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | - | Tris/Glycine buffer without SDS. Compatible with Native-PAGE for proteins and DNA. Enables BN-PAGE of proteins when supplemented with WSE-7067 EzBlueNative Additive. |
| WSE-7056 EzRun ClearNative | - | 〇 | - | - | 〇 Tris-based and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | - | High-resolution Clear Native PAGE running buffer without SDS. Uses an anionic surfactant to minimize electrophoretic interference, allowing efficient separation of native (non-denatured) proteins. |
| WSE-7057 EzRun BlueNative | - | 〇 | - | - | 〇 Tris-based and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | - | Blue Native PAGE running buffer without SDS. Supplied with EzRun BlueNative Additive. Coomassie Brilliant Blue (CBB) enables separation of native protein complexes without dissociation while minimizing electrophoretic interference. |
| WSE-7065 EzRun MOPS | 〇 | - | - | 〇 Bis-Tris–based, imidazole-based, Tris-based, and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | - | Tris/MOPS/SDS buffer compatible with various polyacrylamide gels. Reduces protein mobility, improving resolution in the low–molecular-weight range. High electrophoretic speed enables rapid separation. |
|
| WSE-7066 EzRun MOPS non-SDS | - | 〇 | 〇 | 〇 | 〇 Bis-Tris–based, imidazole-based, Tris-based, and Tris/Glycine–based polyacrylamide gels | 〇 e-PAGEL, e-PAGEL HR, u-PAGEL H, c-PAGEL Neo | 〇 | Tris/MOPS buffer without SDS. Compatible with Native-PAGE for proteins and DNA. Enables BN-PAGE of proteins when supplemented with WSE-7067 EzBlueNative Additive. |
Brochure
Instruction Manual
Specifications
| WSE-7066 EzRun MOPS non-SDS | |
|---|---|
| Components | Tris, MOPS, EDTA |
| Volume | 250 mL (20× concentrate) |
| Usage | Dilute 20-fold with distilled water 220 mL × 22 runs (when using ATTO “Submerge Mini Electrophoresis System”) 450 mL × 11 runs (when using ATTO “Mini Slab Electrophoresis System”) |
| Storage / Shelf Life | 1 year at room temperature, protected from light (unopened) |
Ordering Information
| Code No. | Description | Unit |
|---|---|---|
| 2332306 | WSE-7066 EzRun MOPS non-SDS | 1 pk |